29 Dec Integrated OMICS approach reveals ROCK1 kinase as potential combinatorial drug target in BRAF mutant melanoma
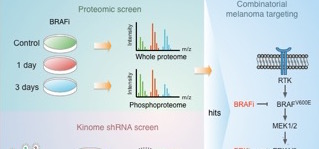
In a recent, open access, article in Molecular Systems Biology researchers from the Biomolecular Mass Spectrometry and Proteomics Group at Utrecht University and The Netherlands Cancer Institute in Amsterdam, report on an integrated analysis of proteomic and phospho‐proteomic data from BRAF inhibitor‐treated melanoma cells and a functional genomic screen for shRNAs sensitizing melanoma to BRAF inhibitor treatment, which identifies ROCK1 kinase as a combinatorial drug target.
Treatment of BRAF mutant melanomas with specific BRAF inhibitors leads to tumor remission. However, most patients eventually relapse due to drug resistance. The researchers designed an integrated strategy using (phospho)proteomic and functional genomic platforms to identify drug targets whose inhibition sensitizes melanoma cells to BRAF inhibition. They found many proteins to be induced upon PLX4720 (BRAF inhibitor) treatment that are known to be involved in BRAF inhibitor resistance, including FOXD3 and ErbB3. Several proteins were down‐regulated, including Rnd3, a negative regulator of ROCK1 kinase.
For the genomic approach, they performed two parallel shRNA screens using a kinome library to identify genes whose inhibition sensitizes to BRAF or ERK inhibitor treatment. By integrating the functional genomic and (phospho)proteomic data, they identified ROCK1 as a potential drug target for BRAF mutant melanoma. ROCK1 silencing increased melanoma cell elimination when combined with BRAF or ERK inhibitor treatment.
Translating this to a preclinical setting, a ROCK inhibitor showed augmented melanoma cell death upon BRAF or ERK inhibition in vitro. This research collaboration provides data that merits exploration of ROCK1 as a target in combination with current BRAF mutant melanoma therapies.
Article
ROCK1 is a potential combinatorial drug target for BRAF mutant melanoma
Marjon A Smit, Gianluca Maddalo, Kylie Greig, Linsey M Raaijmakers, Patricia A Possik, Bas van Breukelen, Salvatore Cappadona, Albert JR Heck, AF Maarten Altelaar, Daniel S Peeper
Molecular Systems Biology(2014)10:772; DOI 10.15252/msb.20145450
Funding
This work was funded by the NWO VIDI grant (723.012.102) awarded to Maarten Altelaar, an International postdoc grant from Vetenskapsrådet (VR), Sweden awarded to Gianluca Maddalo, by the Netherlands Proteomics Center (NPC), the Netherlands Organization for Scientific Research (NWO) funded large-scale proteomics facility Proteins At Work (project 184.032.201), by the European Community’s Seventh Framework Programme (FP7/2007–2013) for the PRIME-XS project grant agreement number 262067, a Queen Wilhelmina award and a grant from KWF Kankerbestrijding
![]()


Sorry, the comment form is closed at this time.